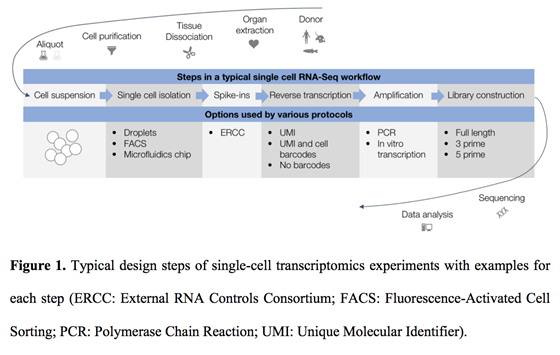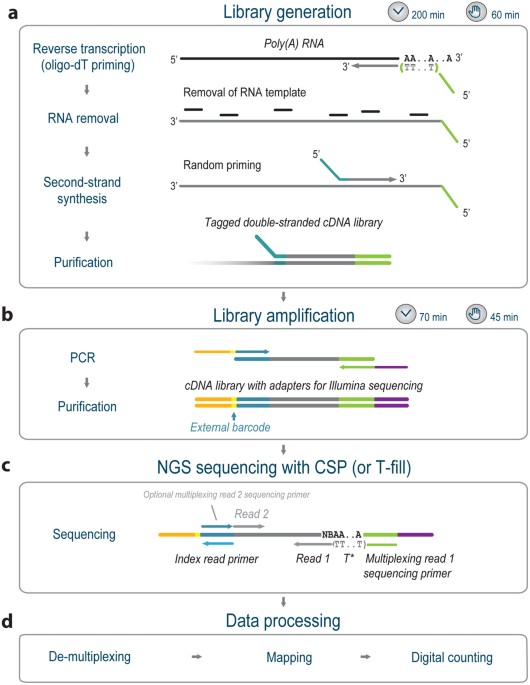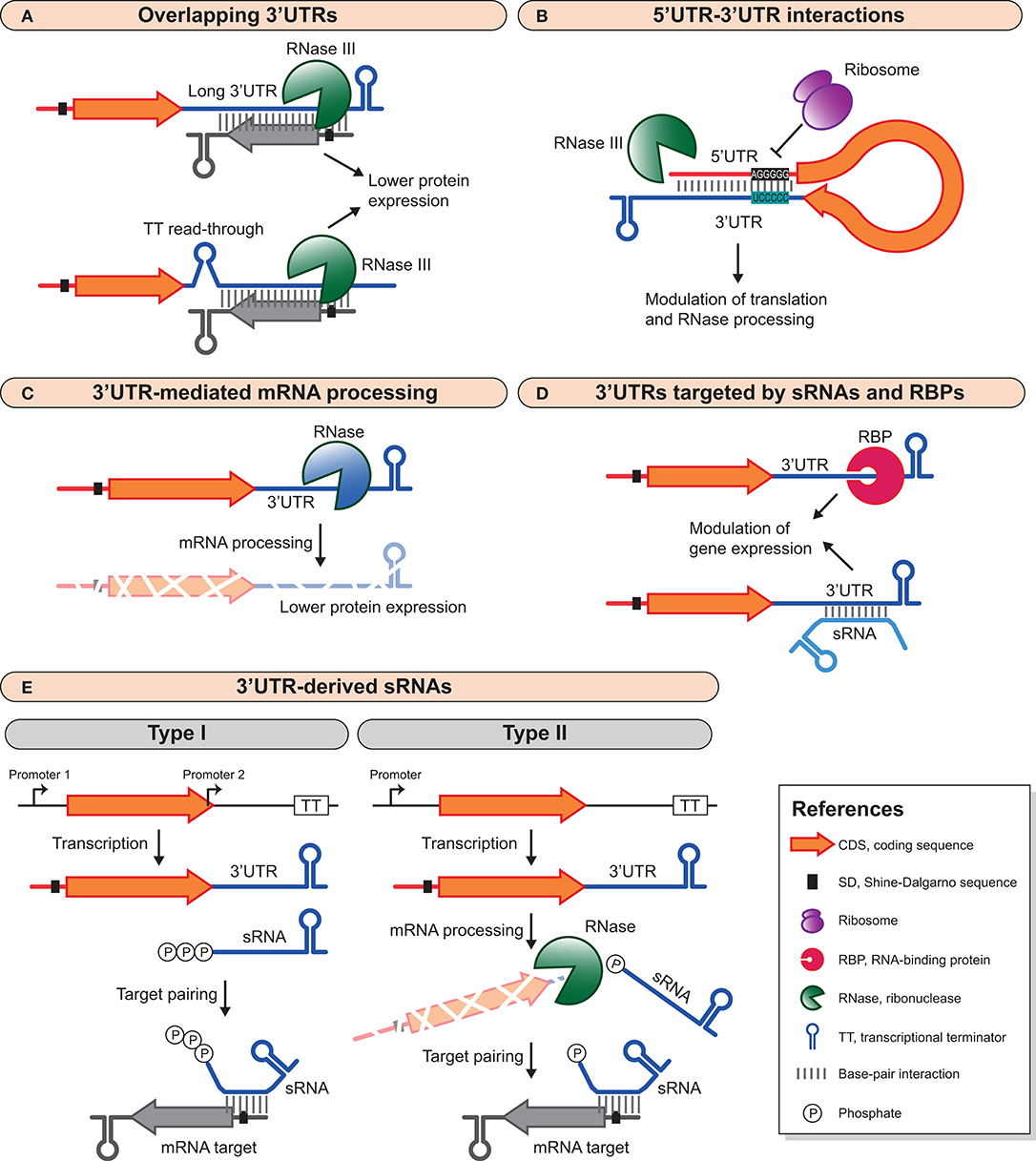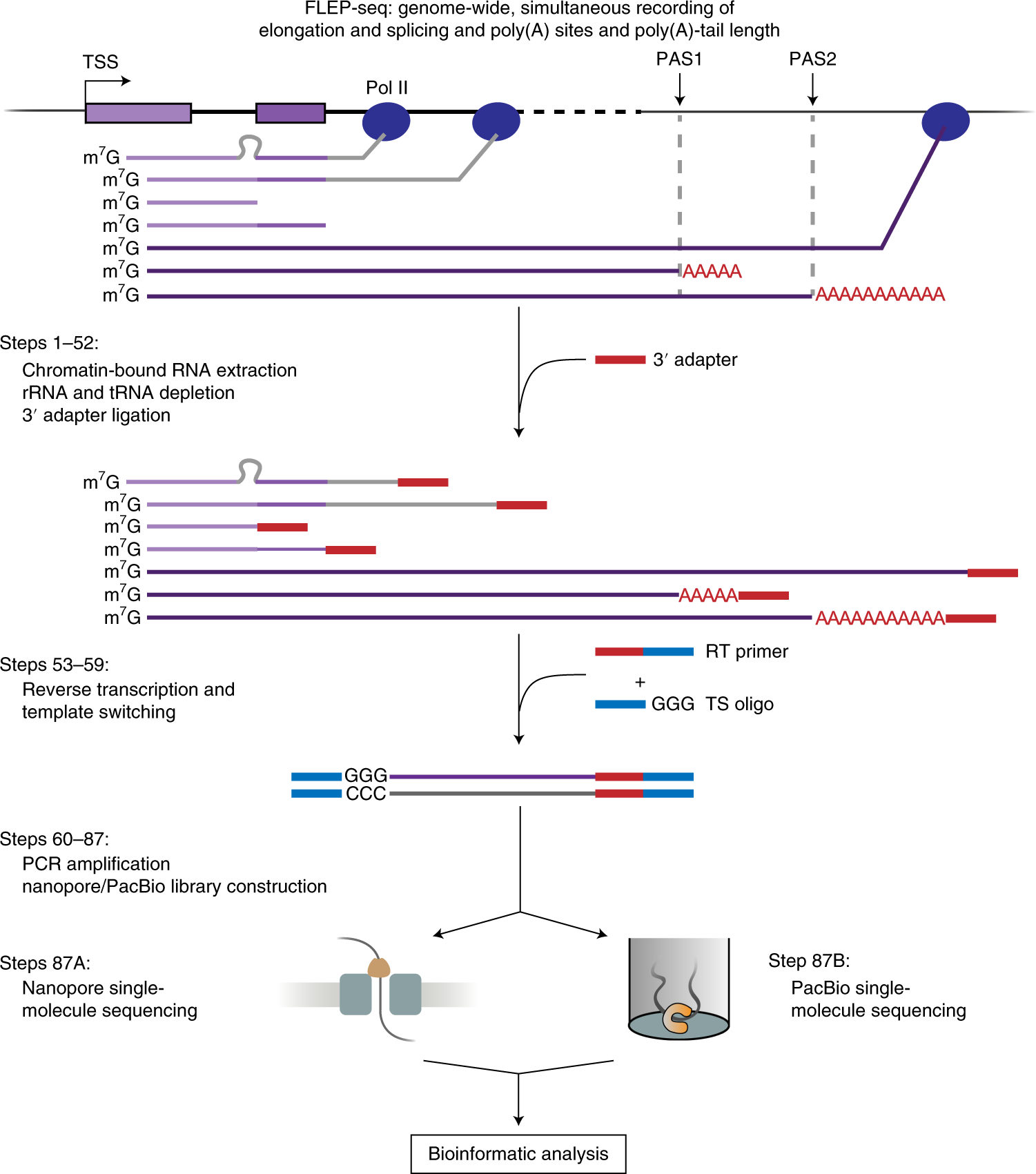
FLEP-seq: simultaneous detection of RNA polymerase II position, splicing status, polyadenylation site and poly(A) tail length at genome-wide scale by single-molecule nascent RNA sequencing | Nature Protocols
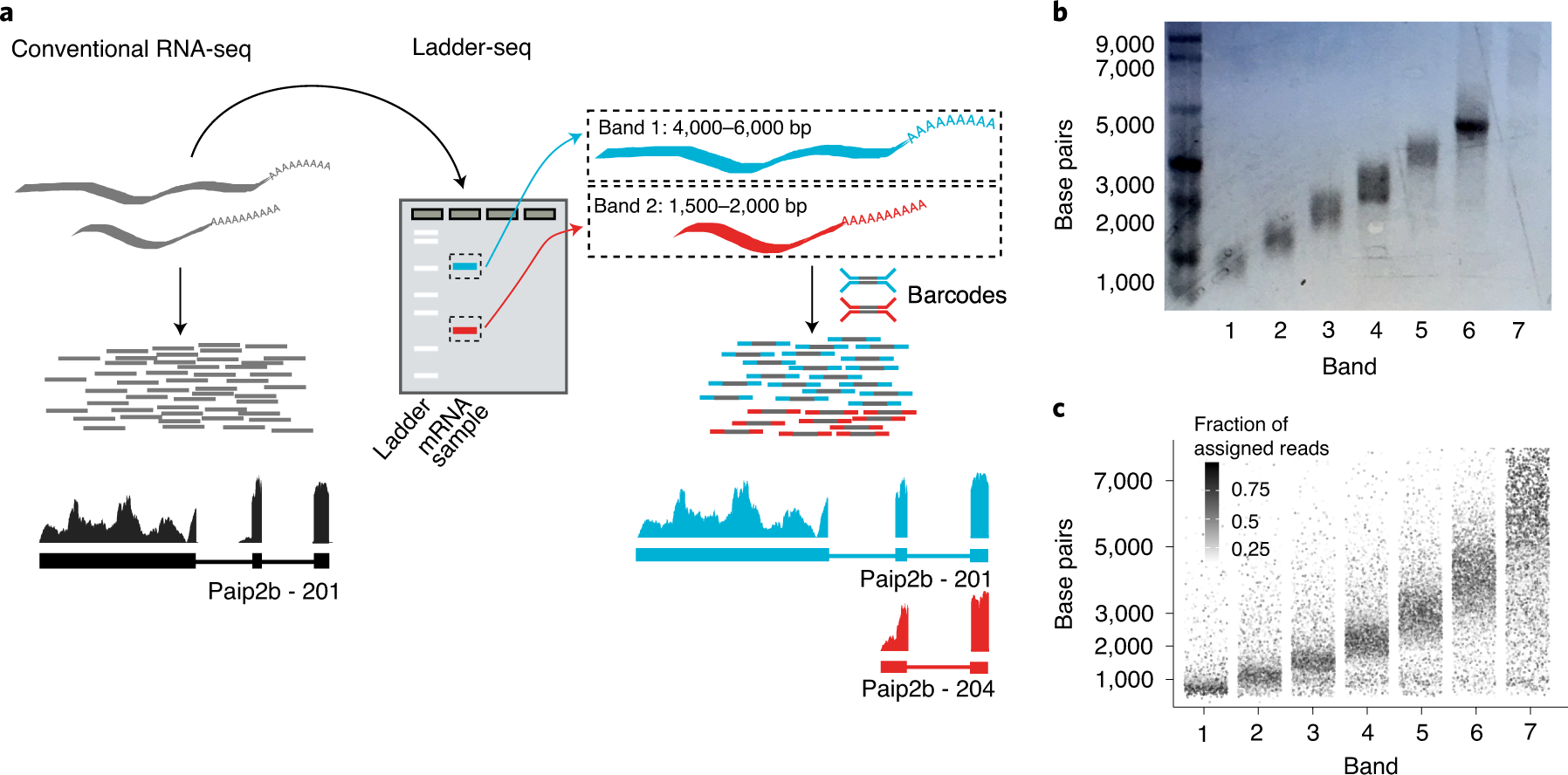
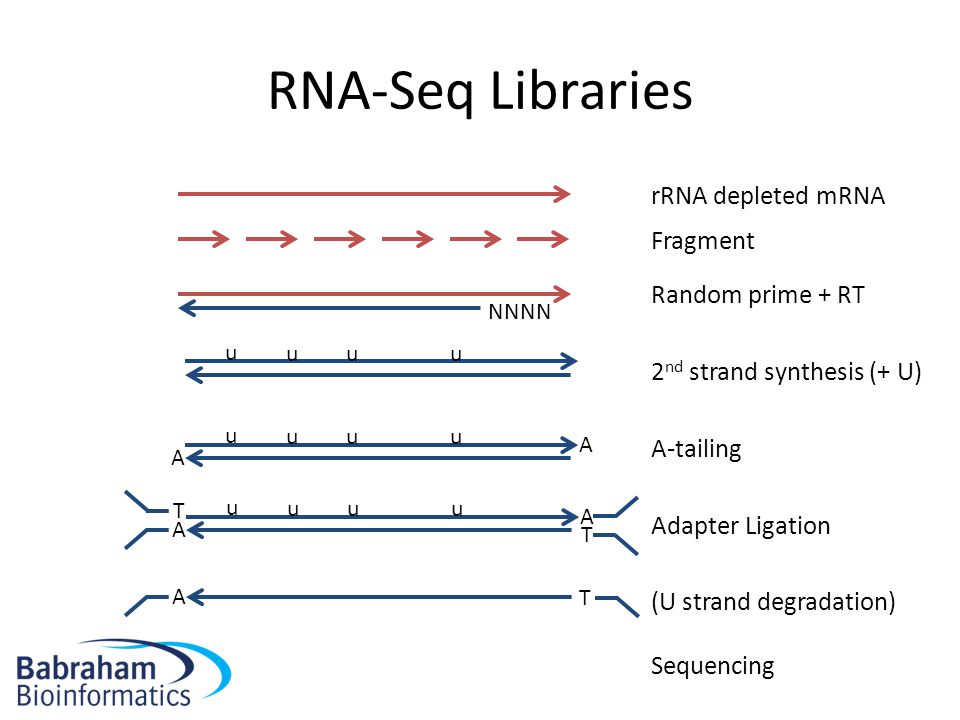
Partitioning RNAs by length improves transcriptome reconstruction from short-read RNA-seq data | Nature Biotechnology
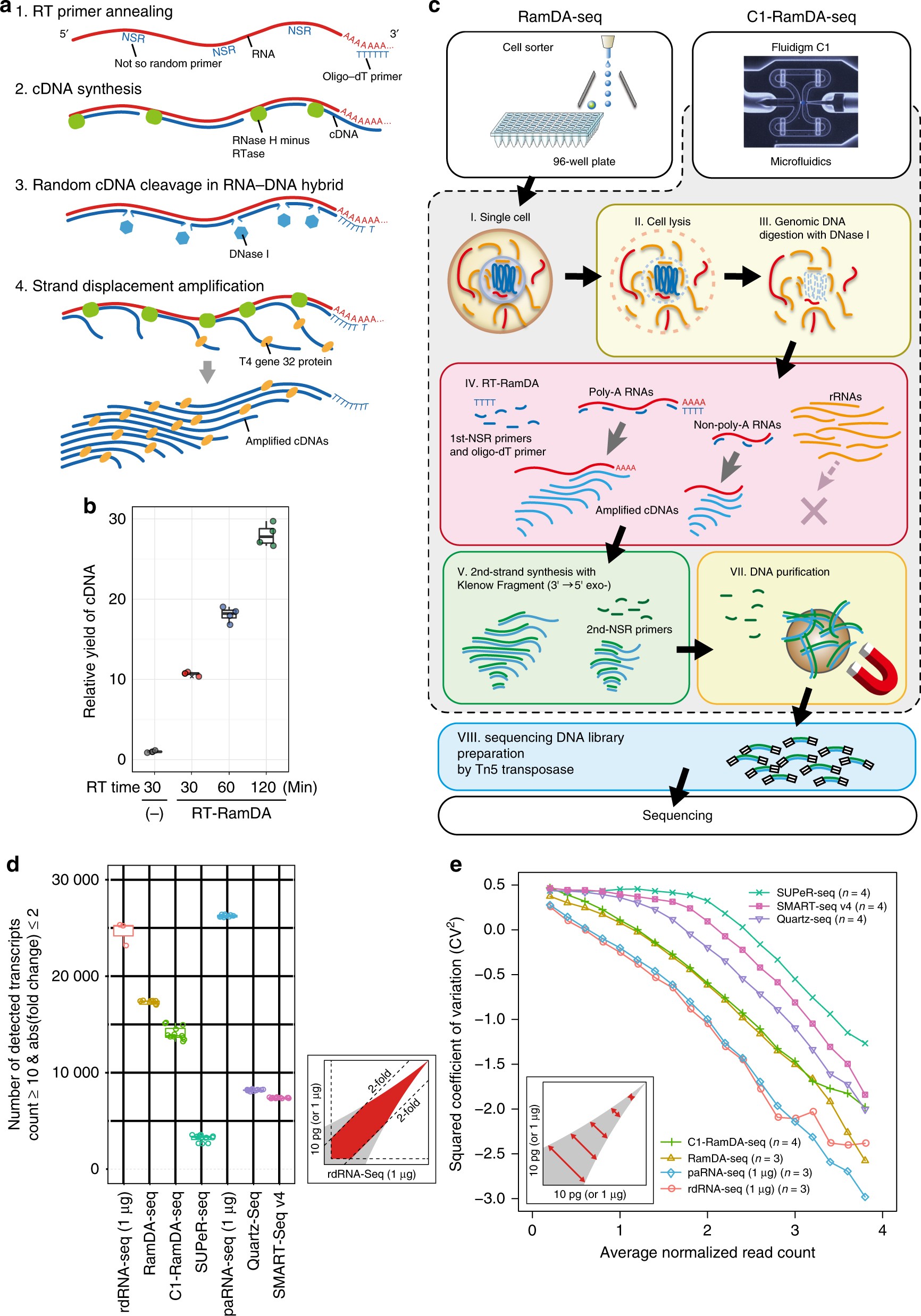
Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs | Nature Communications

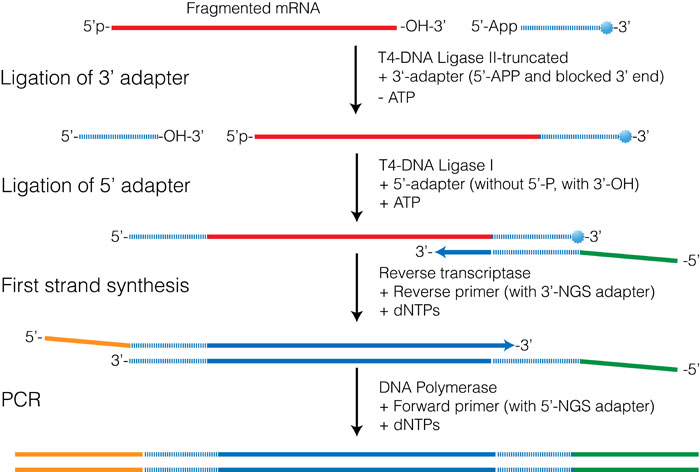
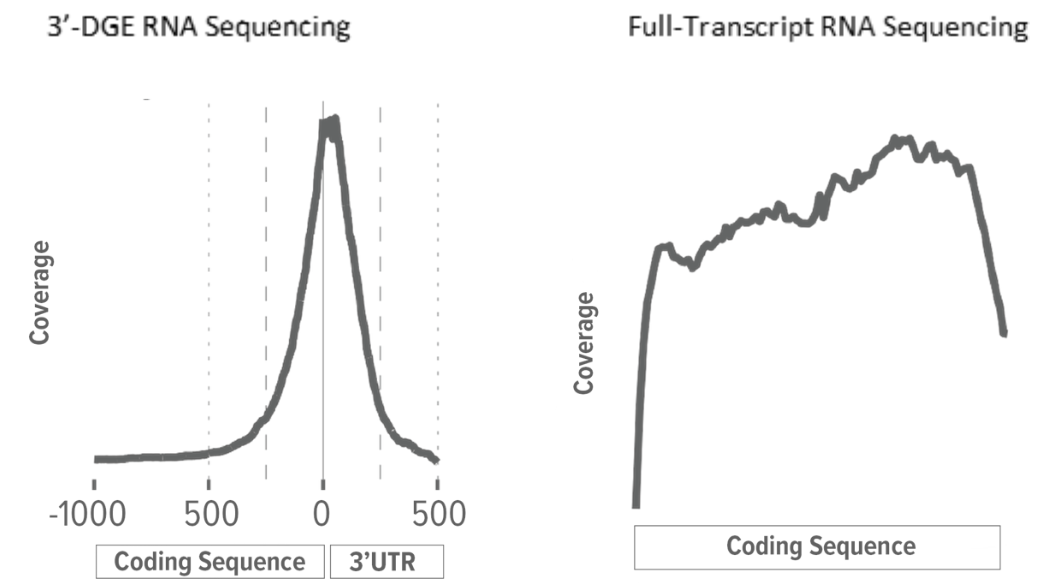
Simultaneous measurement of transcriptional and post-transcriptional parameters by 3′ end RNA-Seq | RNA-Seq Blog

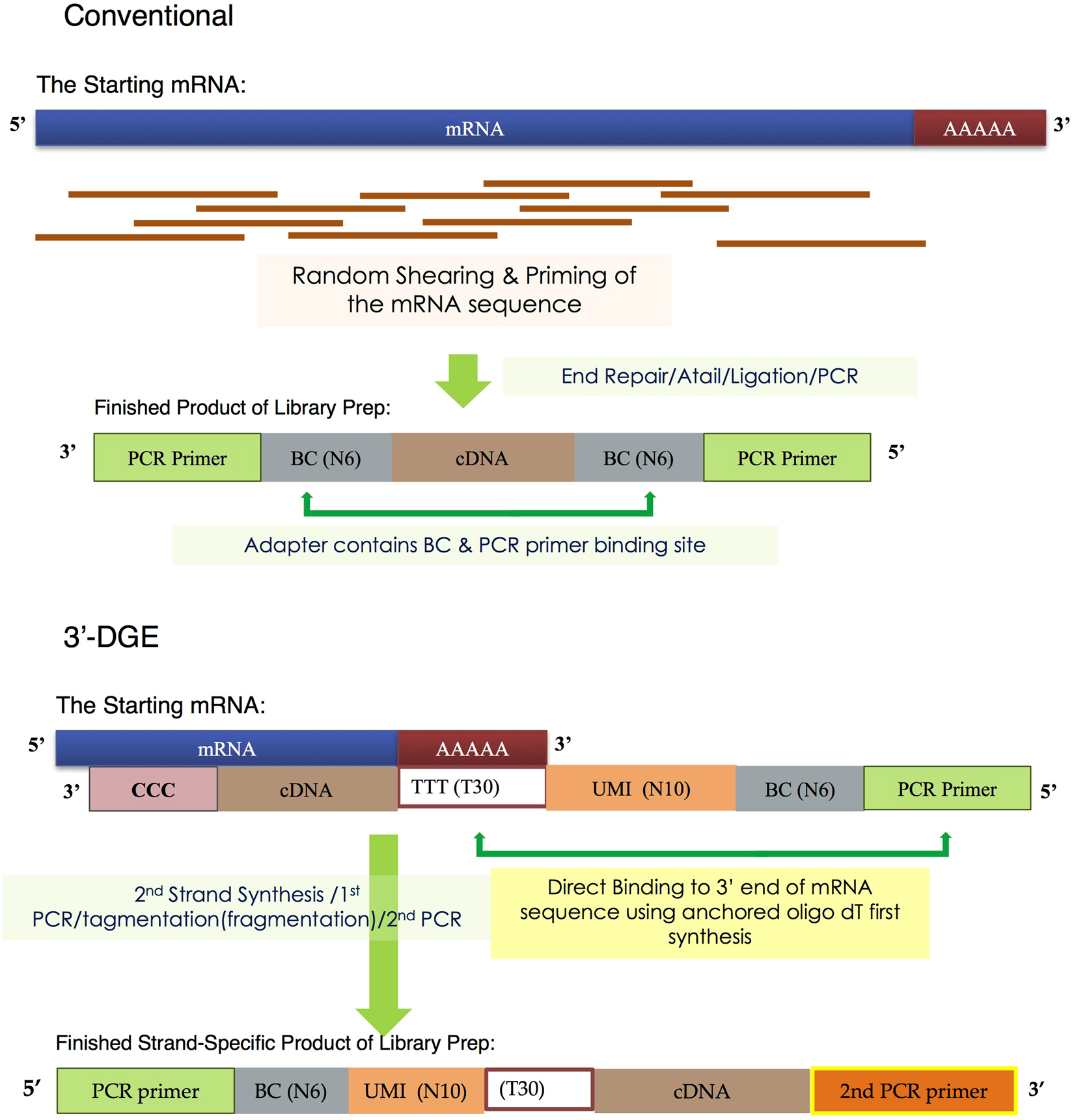
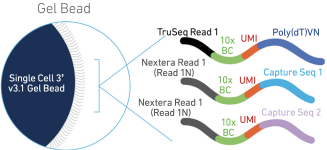
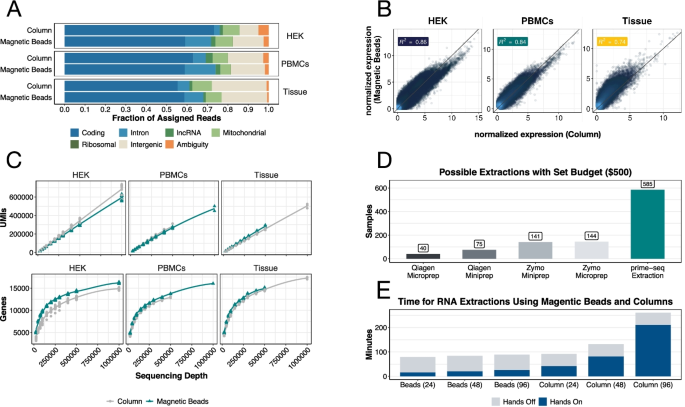
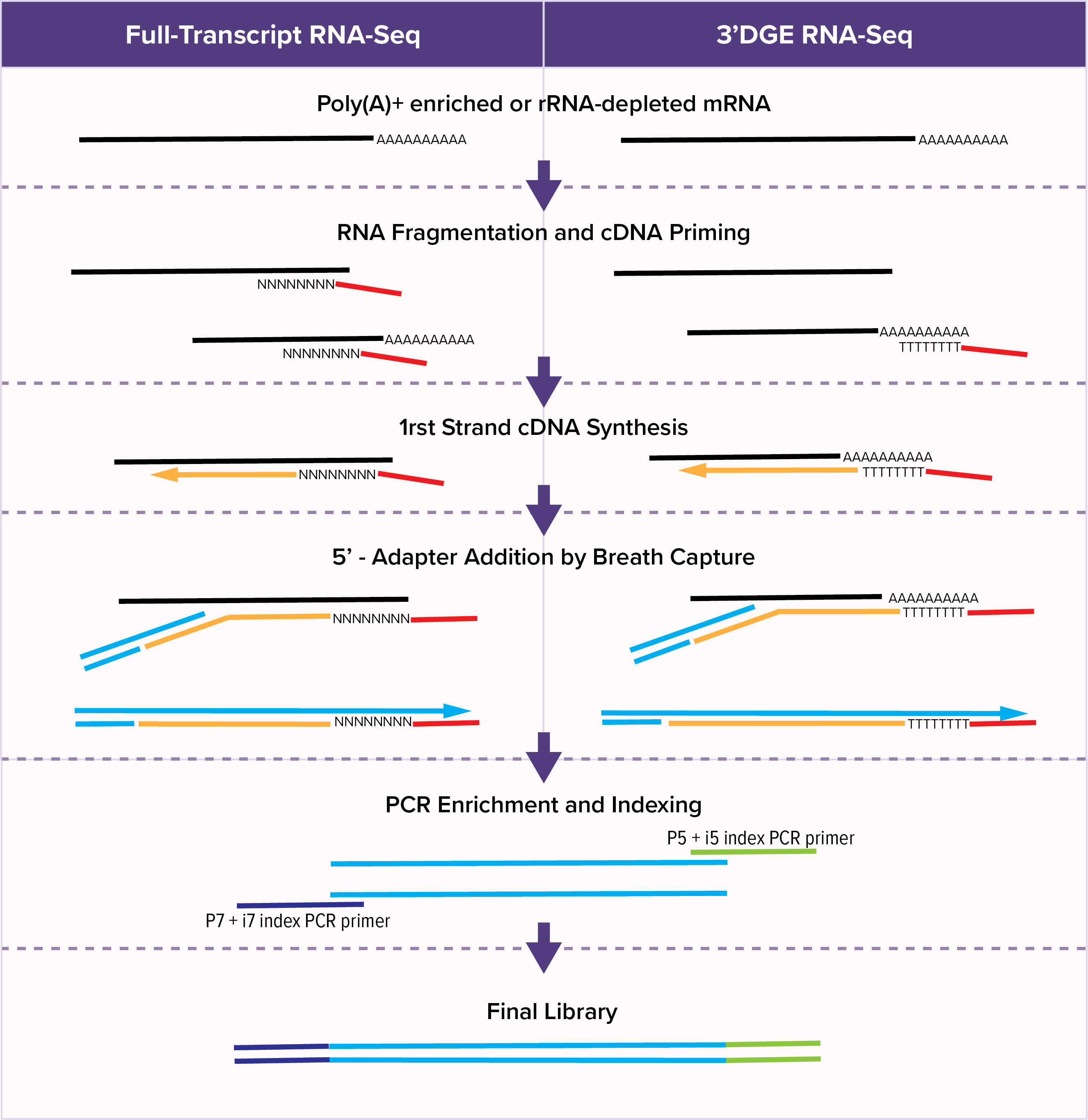
A comparison between whole transcript and 3' RNA sequencing methods using Kapa and Lexogen library preparation methods | BMC Genomics | Full Text